TypOn: The Microbial Typing Ontology
TypOn, the typing ontology, was designed with the goal of have a machine-readable language that describes the field of microbial typing, which is the microbial identification and characterization of strains at subspecies level.
NEWS: TypOn paper is out! Catia Vaz*, Alexandre P Francisco, Mickael Silva, Keith A Jolley, James E Bray, Hannes Pouseele, Joerg Rothganger, Mario Ramirez and Joao A Carrico Typon: the microbial typing ontology was published in Journal of Biomedical Semantics
The present version of TypOn focus only on sequence based typing methods, since, due to the latest developments in DNA sequencing, they have become the standard and most widely methods for bacterial strain identification. Therefore, TypOn is able to describe MLST (Multilocus Sequence Typing) and MLST-like typing schemas, VNTR-based (Variable Number ot Tandem Repeats) methodologies such as MLVA (Multilocus VNTR Analysis) and methodologies based on specific DNA Repeat regions such as Spa typing for Staphylococcus aureus
TypOn ontology design follows a bottom-up approach, were the concepts and relationships were first modeled after the MLST databases , namely pubMLST and MLST.net.
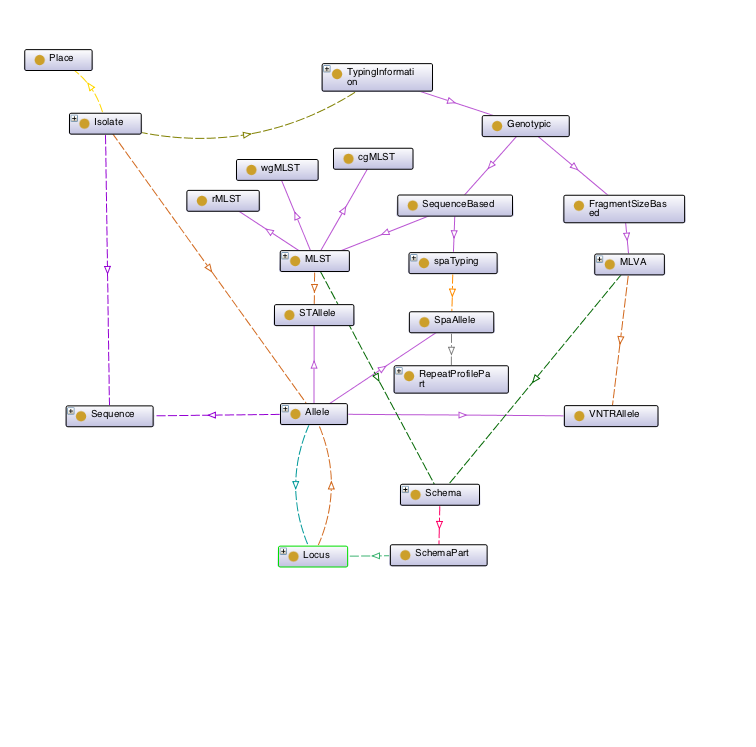
On the left , the image represents the major classes defined by Typon. TypOn was defined in OWL 2.0 with the help of Protégé and Redland RDF Libraries. For more information, and to obtain ontology files, check the development repository on Bitbucket.
The first prototype version of the ontology was published in: Almeida J, Tiple J, Ramirez M, Melo-Cristino J, Vaz C, Francisco AP, Carrico JA. An ontology and a REST API for sequence based microbial typing data. Bioinformatics for Personalized Medicine, Lecture Notes in Bioinformatics. Volume 6620, 2012, pp 21-28 Preprint